Current research in the Imperiali Lab
Life at the membrane frontier
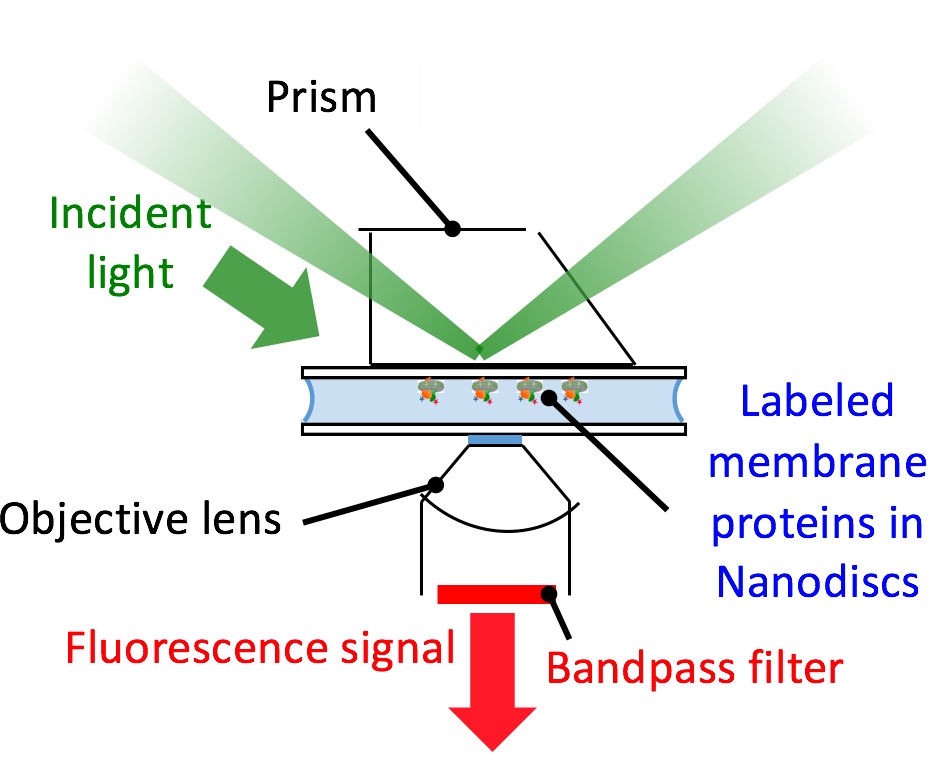
We study the structural and dynamic organization of membrane protein metabolons in model membrane nanoparticles.
To define these supramolecular complexes in native-like environments, we reconstitute protein assemblies in lipid bilayer nanodiscs and styrene-maleic acid nanoparticles and exploit fluorescence-based approaches at the single-molecule and single macromolecular-complex level.
We also investigate how membrane-embedded substrates modulate the biophysical properties of lipid bilayers and how membrane proteins recognize and bind these substrates. We are determining how the membrane architecture of the monotopic superfamily of phosphoglycosyl transferases catalyze phosphosugar transfer to a membrane-embedded polyprenol phosphate acceptor. In our approach, we adapt and integrate synergistic approaches from the fields of chemical biology, biochemistry, biophysics and peptide chemistry to overcome and address the many challenges of studying membrane-associated molecules and macromolecules.
Bioorthogonal protein labeling for biochemistry and biophysics
Detailed studies on intermolecular and intramolecular protein interactions demands selective and efficient protein labeling strategies. We exploit the latest advances in bioorthogonal conjugation including sortase mediated ligation and unnatural amino acid mutagenesis using pyrrolysine tRNA/AARS variants. These two powerful approaches deliver uniquely-modified membrane proteins for detailed biophysical studies.
Analysis of enzyme mechanisms
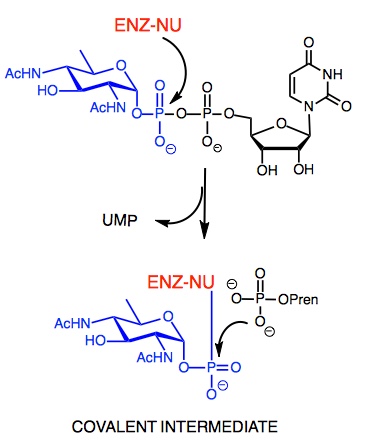
We investigate enzyme mechanisms using a complement of approaches including kinetic analysis, synthesis and investigation of substrate analogs and inhibitors, and identification of trapped, kinetically-relevant intermediates for mass spectrometry and other analytical readouts. These approaches have recently been applied to PglC, a monotopic integral membrane protein that catalyzes phosphoglycosyl transfer in the first committed step of bacterial protein glycosylation.
Medicinal chemistry of antivirulence agents
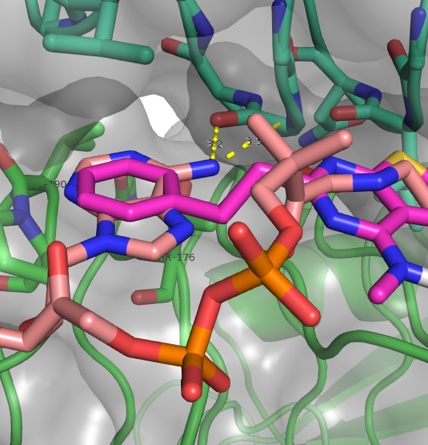
We apply fragment-based screening and structure-driven approaches for development of synthetic small molecule inhibitors of bacterial virulence. The targets that we focus on are the biosynthetic enzymes that lead to prokaryote-specific bacterial sugars commonly found in virulence-associated glycoconjugates. Our synthetic approaches exploit a wide range of reactions, including heterocycle generation, transition metal-catalyzed coupling, and click-type conjugation in order to efficiently synthesize diverse inhibitors for in vitro and in vivo assays as well as structure analysis of inhibitor-bound enzyme complexes.
Natural product mimetics
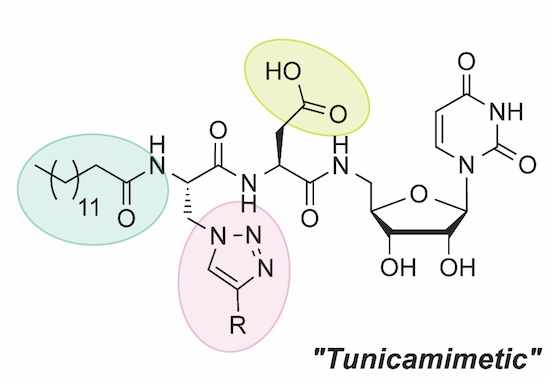
We are inspired by potent natural product inhibitors of glycoconjugate biosynthesis such as mureidomycin and tunicamycin. These compounds share a common uridine scaffold and metal-ion coordinating or displacing groups that interfere with target enzymes. We develop nucleoside-based libraries for targeting glycan biosynthesis and to overcome bottlenecks in traditional synthesis, we develop solid-phase approaches to rapidly access diversity of structures with tunable functions. We then use a newly-established nucleotide-detection assay for high-throughput inhibition studies
Cell-based assays of Campylobacter jejuni pathogenesis
We collaborate with Prof. Linda Griffith in MIT’s Biological Engineering Department to apply chip-based flow devices to study the immune response of human gut intestinal epithelia to the GI pathogen Campylobacter jejuni. The strains we employ are competent or defective in glycosylation, and microbiological and chip-based experiments enable us to evaluate adhesion and invasion into host cells and tissues. In addition to co-culture of human and bacterial cells, we isolate and administer outer membrane vesicles (OMVs) secreted by C. jejuni. OMVs are used for delivery of virulence factors, host invasion, and immune response modulation and our goal is to understand the role that these vesicles play in virulence and to what extent N-linked glycosylation is implicated in C. jejuni pathogenicity.
Reagentless sensors of human biomarkers
In collaboration with Prof. Dane Wittrup in MIT’s Chemical Engineering Department we leverage directed evolution in yeast-surface display to engineer protein scaffolds that bind to disease-relevant biomarkers. The evolved scaffolds, including fibronectin (FN3) or a DNA-binding protein (Sso7d), with desirable ligand-binding properties can be armed with solvatochromic dyes to enable reagentless sensing. These sensors be valuable for reporting and quantitation of diverse analytes including cytokines and other cancer-related biomarkers.
Fluorescent and luminescent tools for biochemistry and biology
Our group develops and implements new organic fluorophores and luminescent lanthanide complexes. Chelation-enhanced and environment-sensitive fluorophores have been introduced and reagents integrating these fluorophores are valuable for protein kinase sensing and the detection of macromolecular interactions respectively. Luminescent peptide-based lanthanide binding tags (LBTs), developed in collaboration with the Allen lab comprise short peptide sequences that are engineered to avidly bind to trivalent lanthanide (Ln3+) ions. LBT sequences impart on fusion proteins a specific lanthanide-binding site for applications that rely upon the unique photophysical properties of the bound complex including advantageous properties for NMR, X-ray crystallography and LRET.